
There are several reasons why this book might be of interest to a TeX user. First, LaTeX has a prominent place in the book. Second, the book describes a very interesting offshoot of literate programming, a topic traditionally popular in the TeX community. Third, since a number of TeX users work with data analysis and statistics, R could be a useful tool for them.
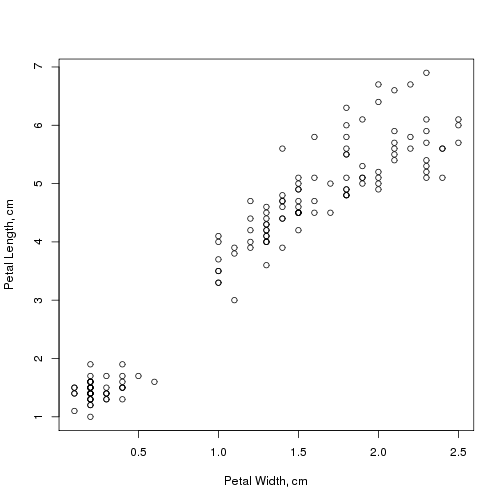
Since some TUGboat readers are likely not familiar with R, I would like to start this review with a short description of the software. R [1] is a free implementation of the S language (sometimes R is called GNU S). The latter is a language for statistical computations created at Bell Labs during its “Golden Age” of computing, when C, awk, Unix, et al. were developed at this famous institution. S is very convenient for a data exploration. For example, consider the dataset iris (included in the base R distribution) containing measurements of 150 flowers of Iris setosa, Iris versicolor and Iris virginica [2]. We may inquire whether petal length and petal width of irises are related. To this end we can plot the data:
plot(Petal.Length ~ Petal.Width, data = iris, xlab = "Petal Width, cm", ylab = "Petal Length, cm")
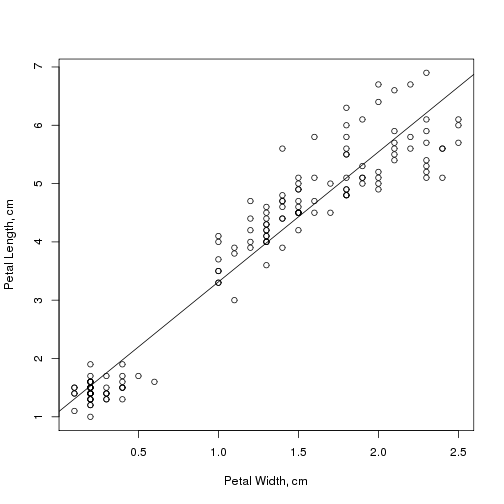
model <- lm(Petal.Length ~ Petal.Width, data = iris)
model$coefficients
## (Intercept) Petal.Width ## 1.084 2.230
summary(model)$r.squared
## [1] 0.9271
plot(Petal.Length ~ Petal.Width, data = iris, xlab = "Petal Width, cm", ylab = "Petal Length, cm") abline(model)
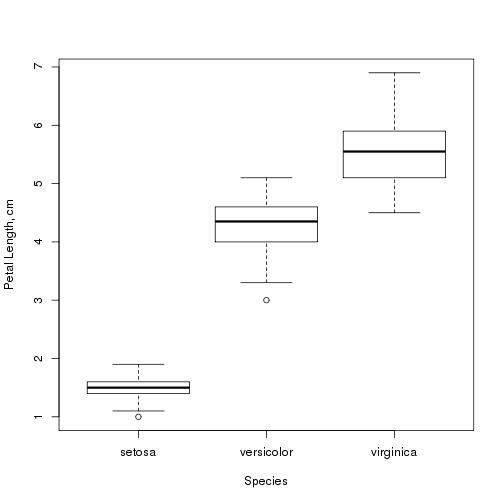
We can also study how petal length depends on the species of iris:
boxplot(Petal.Length ~ Species, data = iris, xlab = "Species", ylab = "Petal Length, cm")
While interactive computations and data exploration are indispensable in such research, one often needs a permanent record that can be stored and shared with other people. A saved transcript of a computer session (or output of a batch job) provides such a record, but in a rather imperfect way. From the transcript one can see what we asked the computer and what the computer replied, but the most important parts of the exploration, namely, why did we ask these questions, which hypotheses were tested, and what our conclusions were, remain outside this record. We can add such information in comments to the R code, but it is awkward to express a complex discussion, often heavy with mathematics, using only an equivalent of an old-fashioned typewriter. This is why many commercial interactive computation systems offer so-called “notebook” interfaces, where calculations are interlaced with the more or less well typeset text and figures. Unfortunately these interfaces, being proprietary, cannot be easily integrated with scientific publishing software, and it takes a considerable effort to translate these “notebooks” into papers and reports. The power of free software is the possibility to combine different building blocks into something new, often not foreseen by their original authors. Thus an idea to combine a free statistical engine and a free typesetting engine is a natural one.
This idea is close to that of literate programming [5]. We have a master document which describes our investigation and contains blocks of text, possibly with equations and figures, and blocks of computational code, that can also generate text, equations and figures. As in the conventional literate programming paradigm, this document can be woven or tangled. Weaving creates a typeset document, while tangling extracts the program (almost) free of comments. However, there is an important difference between the conventional literate programming and the literate programming of data exploration. In the conventional case we are usually interested in the program itself, which is supposed to run many times with different inputs giving different results. For the data exploration the input is usually as important as the program. In most cases we want to run the program just once and show the results. Therefore weaving becomes a more complex process, involving running the tangled program and inserting the results in the appropriate places of the typeset document. On the other hand, tangling by itself is used more rarely (but is still useful in some cases, for example, to typeset this review, as described below).
The first (and a very successful) attempt to apply the ideas of literate programming to S was the package Sweave [6]. Uwe Ziegenhagen introduced the package to the TeX community in his brilliant talk at TUG 2010 [7]. In Sweave we write a file source.rnw, which looks like a TeX file, but includes special fragments between the markers <<...>> and @ (this notation is borrowed from the Noweb literate programming system [8]). For example, the box plot above can be produced by the following fragment of an .rnw file:
We also can “inline” R code using the command \Sexpr, for example,
Note the different usage of $ inside \Sexpr (a part of R code) and outside it (TeX math mode delimiter).
When we feed the file source.rnw to Sweave, it runs the R code in the chunks, and replaces these chunks and \Sexpr macros by the properly formatted results and/or the code itself. This produces a “weaved” file source.tex. We can control the process by the options inside <<...>>. For example, setting there echo=FALSE, we suppress the “echoing” of the source code. We can wrap a fragment into a figure environment and get a result like the one in Figure 1. Alternatively we can use R functions that output LaTeX code and, setting the proper options, get a table like Table 1.
These figures and tables, as well as inline expressions, can be configured to hide the code, for example for a formal publication. One can write a paper as an .rnw file and submit a PDF or TeX file to a journal without copying and pasting the results of the calculations, eliminating the risk of introducing new errors and typos. The versatility of this approach allows one to create quite varied publications, from a conference poster to a book. On the other hand, if the user does not suppress the code, a nicely formatted and reproducible lab report is produced.
Since Sweave was written, many packages have been devised to extend its capabilities and to add new features to it. At some point the user community felt the need for a refactoring of Sweave with better integration of the new features and a more modular design. The package knitr [9], which lists Yihui Xie as one of its principal authors, provides such refactoring. The name “knitr” is a play on “Sweave”. The knitr functions performing weaving and tangling are called “knit” and “purl” correspondingly.
| Parameter | Est. | σ | t | p-value
|
| Intercept | 1.08 | 0.0730 | 14.8 | 4.04e-31 |
| Slope | 2.23 | 0.0514 | 43.4 | 4.68e-86 |
An important feature of knitr is the closeness of its syntax to that of Sweave. Up to version 1.0 knitr supported full compatibility with Sweave, but even today most Sweave code runs without problems in knitr (the function Sweave2knitr can help in the remaining cases). For example, the code producing the iris boxplot above becomes the following in knitr:
The only difference between this code and Sweave code is the absence of fig=TRUE option, which is not needed for the new package. However, the Sweave code above still works in knitr. This makes the switch to the new package rather easy.
It should be noted that some of the features of knitr discussed below are also available in Sweave when using add-on packages; knitr offers them “out of the box” and better integrates them.
For example, knitr has two dozen or so different graphics formats (or “devices”) to save the graphics. One very interesting device is tikz, which can be used to add TeX annotations to the plots.
Another useful feature of knitr is the option of caching the computation results. R calculations can often take a significant time. The full recompilation of all chunks after a mere wording change in the TeX part may be too slow for a user. This problem is addressed by the options cache and dependson in knitr. They instruct R to recalculate only the modified chunks and the chunks that depend on them (a clever hashing of the code is used to determine whether a chunk was modified between the runs).
The typesetting of the input code in knitr is closer to the requirements of literate programming than the simple Sweave output: knitr can recognize R language elements like keywords, comments, variables, strings etc., and highlight them according to user’s specifications.
While these features are nice, the real selling points of knitr are its flexibility and modularization. The package has many options for fine-tuning the output. The modular design of the package makes adding new options just a matter of writing a new “hook” (an R function called when processing a chunk). Since the chunk headers in knitr, unlike Sweave, are evaluated as R expressions, one can write quite sophisticated “hooks”.
The modularity of knitr allowed the authors to introduce new typesetting engines. Besides LaTeX, the package can work with other markup languages, e. g. HTML, Markdown and reStructuredText.
The flexibility and ease of customization of knitr are especially useful for book publishing. Yihui Xie lists several books created with knitr. Barbara Beeton [10] reports a positive experience with AMS publishing such books.
This review turned out to be a test of the flexibility of knitr. At this point an astute reader may have already guessed that it was written as an .rnw file. However, that is not the full story. TUGboat reviews are published in the journal, and the HTML versions are posted on the web at http://www.tug.org/books. As a matter of policy, the HTML version is automatically generated from the same source as the hard copy. This created a certain challenge for this review. First, the hard copy uses plots in PDF format, while the Web version uses them in PNG format. Second, due to the differences in column widths R code should be formatted differently for the print and the Web versions. Third, code highlighting of R chunks was done using font weights for the print version and using colors for the Web version. The following work flow was used: (1) The review was written as an .rnw file. (2) A .tex file was knit and an .R file was purled from the .rnw source. (3) The .tex file was copy-edited by the TUGboat editors. (4) A .pdf output for the journal was produced from this .tex file. (5) A special .Rhtml file was produced from the same .tex file with tex4ht. This file included commands that read the .R program and inserted the code chunks in the proper positions. (6) This .Rhtml file was knit again to produce HTML output and the images for the Web. All this but the actual writing and copy-editing was done by scripts without human intervention.
The package knitr is being actively developed, and many new features are being added. I would like to mention a feature that I miss in the package. The default graphics device, PDF, uses fonts different from the fonts of the main document, and does not allow TeX on the plots. While the tikz device is free of this limitation, it is very slow and strains TeX memory capacity when producing large plots. I think the trick used by the Gnuplot pslatex terminal [11] might be very useful. This terminal creates two files: a TeX file with the textual material put in the proper places using the picture environment, and a graphics file with the graphical material, included through the \includegraphics command. This terminal is much faster than tikz and more flexible than pdf. Moreover, since the TeX file is evaluated in the context of the main document, one can include \ref and other LaTeX commands in the textual part.
To use knitr on the most basic level it is enough to know two simple rules and one option: (1) put R code between <<...>> and @; (2) use \Sexpr{code} for inline R code; (3) use echo=FALSE to suppress echoing the input if necessary. However, since knitr has many options and is highly customizable, one might want to learn more about it to use it efficiently. This justifies the existence of books like the one by Yihui Xie. While there are many free sources of information about knitr, including its manual and the author’s site (http://yihui.name/knitr), there are important reasons why a user would consider investing about $60 in the book.
The book provides a systematic description of the package, including its concepts, design principles, and philosophy. It also has many examples, well thought out advice, and useful tips and tricks.
Here is just one of the techniques I learned from the book. There are two ways of presenting calculation results: using the option echo=TRUE (default) we typeset R code, while using the option echo=FALSE we suppress it. The inclusion of the code has both advantages and disadvantages: we tell the reader more with the code included, but we risk overwhelming and confusing the audience with too much detail. One way to solve this problem is to put only the results in the main text, and show the code itself in an appendix. Of course we do not want to copy and paste the same code twice; a computer can take care of this much better. The book describes how to make this automatic by putting in the appendix these lines:
There are many other useful tips on the pages of this book. Some of them can be found at http://yihui.name/knitr/ and http://tex.stackexchange.com/. Having all these tips collected in a book saves time, however, and reading the book helps to learn knitr and the general interaction of TeX and R.
The book is well written. It has introductory material useful for novices as well as advice for more seasoned users, all explained in conversational English without unnecessary technical jargon. The book describes several integrated work environments (RStudio, LY X, Emacs/ESS) and the interaction of knitr with popular LaTeX packages such as beamer and listings. It discusses in depth package options for formatting input and output, graphics, caching, code reuse, cross-referencing and other purposes. Besides “traditional” applications such as publishing books and reports, the author describes the use of knitr for writing blog entries, student homework, etc. The book covers inclusion of fragments of code written in languages other than R (Python, Perl, C/C++, etc.), dynamic plots with the animate LaTeX package, macro preprocessing and many other topics.
The book itself is written, of course, in knitr and LaTeX (with LY X as the integrated environment). It is well typeset and nicely printed. Regrettably, I find its index rather inadequate: it has only two pages and omits many key concepts. A book like this should at a minimum have an alphabetic list of all package options. However, other than this, I like the way the book is written and published.
While I have been using Sweave and then knitr for several years, I still learned many new useful things from the book. Thus I think it is worth investing money and reading time.
I think the book deserves a place on the bookshelves of both new and experienced R and TeX users.
[1] R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria, 2013. ISBN 3-900051-07-0.
[2] Edgar Anderson. The irises of the Gaspe Peninsula. Bulletin of the American Iris Society, 59:2–5, 1935.
[3] Peter Dalgaard. Introductory Statistics with R. Statistics and Computing. Springer, New York, second edition, 2008.
[4] W. N. Venables and B. D. Ripley. Modern Applied Statistics with S. Statistics and Computing. Springer, New York, fourth edition, 2010.
[5] Donald Ervin Knuth. Literate Programming. Number 27 in CSLI lecture notes. Center for the Study of Language and Information, Stanford, CA, 1992.
[6] Friedrich Leisch. Sweave: Dynamic generation of statistical reports using literate data analysis. In Wolfgang Härdle and Bernd Rönz, editors, Compstat 2002 — Proceedings in Computational Statistics, pages 575–580. Physica Verlag, Heidelberg, 2002. ISBN 3-7908-1517-9.
[7] Uwe Ziegenhagen. Dynamic reporting with R/Sweave and LaTeX. TUGboat, 31(2):189–192, 2010. http://tug.org/TUGboat/tb31-2/tb98ziegenhagen.pdf.
[8] Norman Ramsey. Literate programming simplified. IEEE Software, 11(5):97–105, 1994. http://www.cs.tufts.edu/~nr/noweb/.
[9] Alastair Andrew, Alex Zvoleff, Brian Diggs, Cassio Pereira, Hadley Wickham, Heewon Jeon, Jeff Arnold, Jeremy Stephens, Jim Hester, Joe Cheng, Jonathan Keane, J.J. Allaire, Johan Toloe, Kohske Takahashi, Michel Kuhlmann, Nacho Caballero, Nick Salkowski, Noam Ross, Ramnath Vaidyanathan, Richard Cotton, Romain Francois, Sietse Brouwer, Simon de Bernard, Taiyun Wei, Thibaut Lamadon, Tom Torsney-Weir, Trevor Davis, Weicheng Zhu, Wush Wu, and Yihui Xie. knitr: A general-purpose package for dynamic report generation in R, 2013. R package version 1.5.
[10] Barbara Beeton. Private communication, 2014.
[11] Philipp K. Janert. Gnuplot in Action. Understanding Data with Graphs. Manning Publications Co., 2009.